ERG protein expression and gene rearrangements are present at lower rates in metastatic and locally advanced castration-resistant prostate cancer compared to localized disease.
Abstract
OBJECTIVE:
To compare ERG expression and gene rearrangements rates in metastatic and castration–resistant prostate cancer (CRPC) to localized disease as ERG is the most common genetic event in early prostate cancer (PCa) with potential prognostic and therapeutic implications.
METHODS:
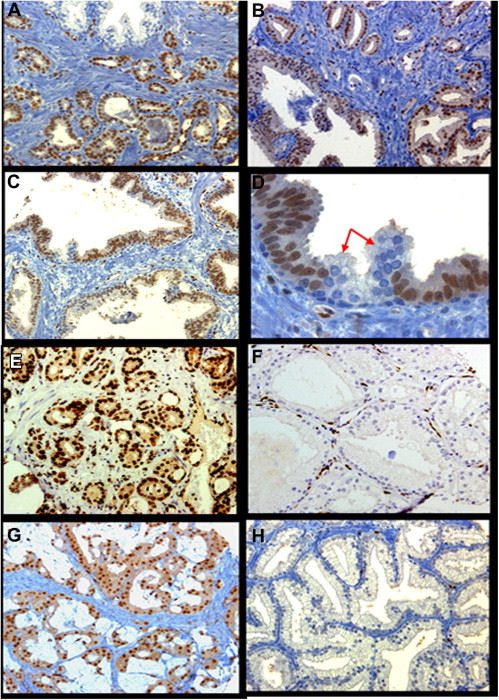
We evaluated ERG protein expression in 344 patients with PCa in 3 cohorts including localized, metastatic, and castration–resistant disease using immunohistochemistry (IHC) and fluorescence in situ hybridization (FISH).
RESULTS:
ERG protein expression was detected exclusively in the neoplastic epithelium and was found in 6.8% and 46.3% of high-grade prostatic intraepithelial neoplasia (HGPIN) and localized PCa, respectively. In metastatic and locally advanced CRPC, ERG expression was significantly lower, occurring at 36.1% and 37.2%, respectively. In PCa with foamy gland morphology, ERG protein expression was detected in only 18.6% compared with reported rates of about 42%-48% in acinar PCa. Moreover, ERG protein expression and gene rearrangements showed an overall consistency rate of 90.6% (P <.0001). The consistency rate was 100% both in benign glands and HGPIN, and 96.1% in localized PCa. However, it was significantly lower at 76.9% and 85% in node metastatic and CRPC, respectively (P <.0001).
CONCLUSION:
ERG protein expression is restricted to neoplastic prostatic epithelium and is present at lower rates in metastatic and CRPC compared to localized PCa. IHC and FISH concordance rates were significantly lower in node metastatic and CRPC compared to localized PCa, which may suggest different biological and therapeutic implications. The lower rate of ERG protein expression in foamy gland PCa may suggest potential differences for this pattern of PCa at the molecular level.
